Cell subsampling using TopACeDo¶
[1]:
%load_ext autotime
%config InlineBackend.figure_format = 'retina'
import scarf
scarf.__version__
[1]:
'0.10.1'
time: 994 ms (started: 2021-08-22 18:56:41 +00:00)
1) Installing dependencies¶
We need to install the TopACeDo algorithm to perform subsampling:
[2]:
!pip install git+https://github.com/fraenkel-lab/pcst_fast.git@deb3236cc26ee9fee77d5af40fac3f12bb753850
!pip install -U topacedo
Collecting git+https://github.com/fraenkel-lab/pcst_fast.git@deb3236cc26ee9fee77d5af40fac3f12bb753850
Cloning https://github.com/fraenkel-lab/pcst_fast.git (to revision deb3236cc26ee9fee77d5af40fac3f12bb753850) to /tmp/pip-req-build-whvvm9rx
Running command git clone -q https://github.com/fraenkel-lab/pcst_fast.git /tmp/pip-req-build-whvvm9rx
Running command git rev-parse -q --verify 'sha^deb3236cc26ee9fee77d5af40fac3f12bb753850'
Running command git fetch -q https://github.com/fraenkel-lab/pcst_fast.git deb3236cc26ee9fee77d5af40fac3f12bb753850
Running command git checkout -q deb3236cc26ee9fee77d5af40fac3f12bb753850
Resolved https://github.com/fraenkel-lab/pcst_fast.git to commit deb3236cc26ee9fee77d5af40fac3f12bb753850
Requirement already satisfied: pybind11>=2.1.0 in /home/docs/checkouts/readthedocs.org/user_builds/scarf/envs/0.10.1/lib/python3.8/site-packages (from pcst-fast==1.0.7) (2.7.1)
Building wheels for collected packages: pcst-fast
Building wheel for pcst-fast (setup.py) ... - \ | / - \ done
Created wheel for pcst-fast: filename=pcst_fast-1.0.7-cp38-cp38-linux_x86_64.whl size=840717 sha256=efe0f4e31902a2de76299ddf9b38a0d4b50522f6ea0da499f6e2948d7b729e4b
Stored in directory: /home/docs/.cache/pip/wheels/48/f1/74/6bb2e9cc262228dbdc63916ec60f0717574b5f633b6ee30b0a
Successfully built pcst-fast
Installing collected packages: pcst-fast
Successfully installed pcst-fast-1.0.7
Requirement already satisfied: topacedo in /home/docs/checkouts/readthedocs.org/user_builds/scarf/envs/0.10.1/lib/python3.8/site-packages (0.2.7)
time: 13 s (started: 2021-08-22 18:56:42 +00:00)
2) Fetching pre-processed data¶
[3]:
# Loading preanalyzed dataset that was processed in the `basic_tutorial` vignette
scarf.fetch_dataset('tenx_5K_pbmc_rnaseq', as_zarr=True, save_path='scarf_datasets')
INFO: Download finished! File saved here: /home/docs/checkouts/readthedocs.org/user_builds/scarf/checkouts/0.10.1/docs/source/vignettes/scarf_datasets/tenx_5K_pbmc_rnaseq/data.zarr.tar.gz
INFO: Extracting Zarr file for tenx_5K_pbmc_rnaseq
time: 15.8 s (started: 2021-08-22 18:56:55 +00:00)
[4]:
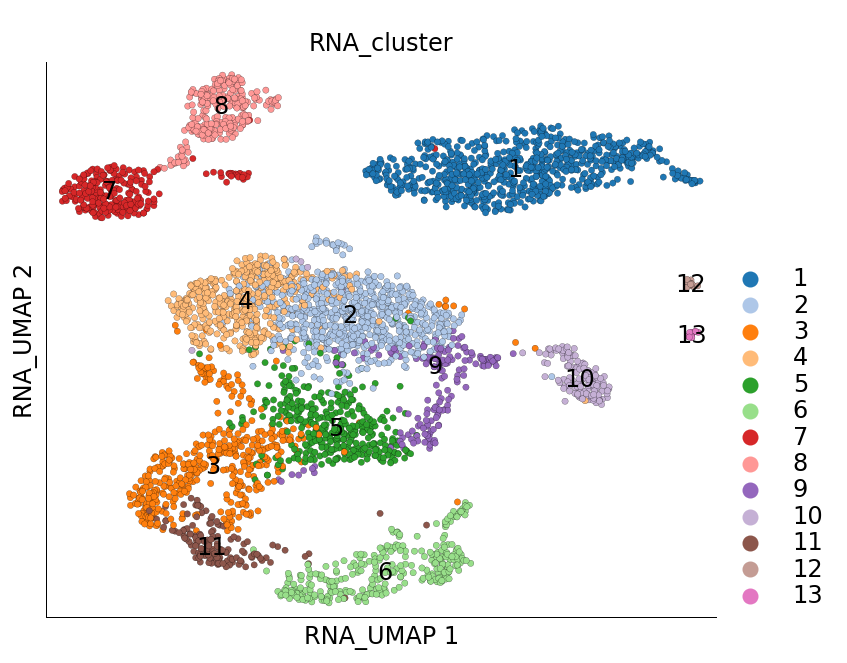
ds = scarf.DataStore('scarf_datasets/tenx_5K_pbmc_rnaseq/data.zarr')
ds.plot_layout(layout_key='RNA_UMAP', color_by='RNA_cluster')
time: 1.09 s (started: 2021-08-22 18:57:11 +00:00)
3) Run TopACeDo downsampler¶
UMAP, clustering and marker identification together allow a good understanding of cellular diversity. However, one can still choose from a plethora of other analysis on the data. For example, identification of cell differentiation trajectories. One of the major challenges to run these analysis could be the size of the data. Scarf performs a topology conserving downsampling of the data based on the cell neighbourhood graph. This downsampling aims to maximize the heterogeneity while sampling cells from the data.
Here we run the TopACeDo downsampling algorithm that leverages Scarf’s KNN graph to perform a manifold preserving subsampling of cells. The subsampler can be invoked directly from Scarf’s DataStore object.
[5]:
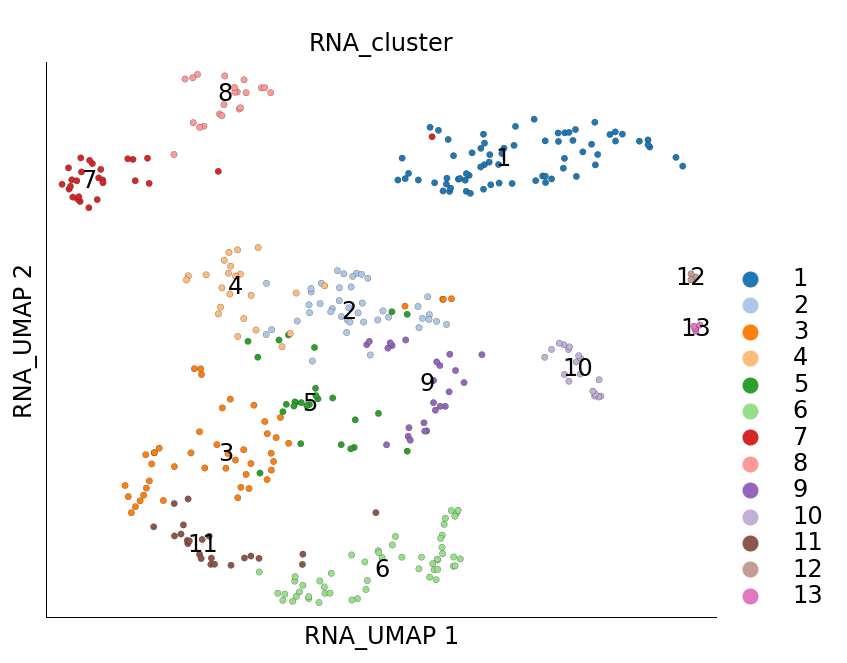
ds.run_topacedo_sampler(cluster_key='RNA_cluster', max_sampling_rate=0.1)
Constructing graph from dendrogram: 100%|██████████| 3939/3939 [00:00<00:00, 23602.32it/s]
INFO: 384 cells (9.75%) sub-sampled. Subsample to Seed (165 cells) ratio: 2.327
time: 1.35 s (started: 2021-08-22 18:57:12 +00:00)
As a result of subsampling the subsampled cells are marked True under the cell metadata column RNA_sketched. We can visualize these cells using plot_layout
[6]:
ds.plot_layout(layout_key='RNA_UMAP', color_by='RNA_cluster', subselection_key='RNA_sketched')
time: 478 ms (started: 2021-08-22 18:57:13 +00:00)
It may also be interesting to visualize the cells that were marked as seed cells used when PCST was run. These cells are marked under the column RNA_sketch_seeds.
[7]:
ds.plot_layout(layout_key='RNA_UMAP', color_by='RNA_cluster', subselection_key='RNA_sketch_seeds')

time: 464 ms (started: 2021-08-22 18:57:14 +00:00)
4) Intermediate parameters of downsampling¶
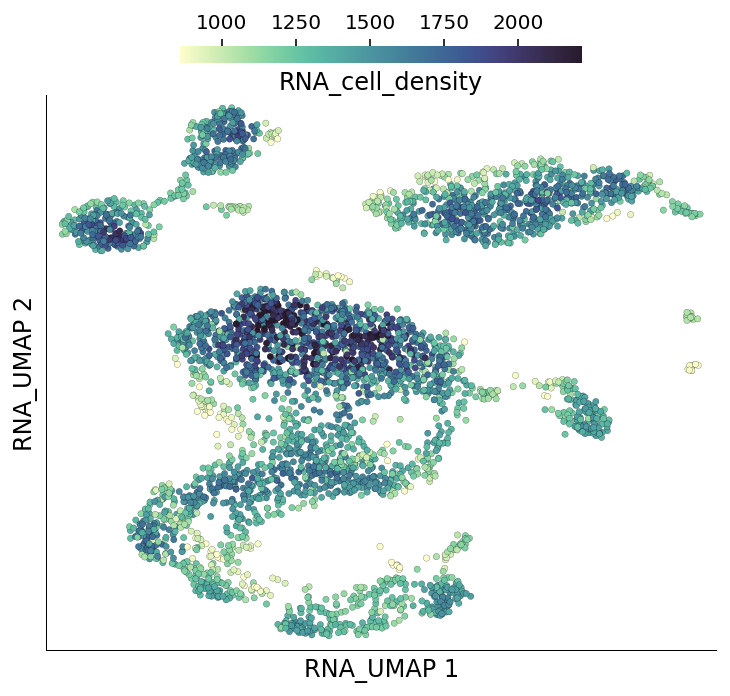
To identify the seed cells, the subsampling algorithm calculates cell densities based on neighbourhood degrees. Regions of higher cell density get a sampling penalty. The neighbourhood degree of individual cells are stored under the column RNA_cell_density.
[8]:
ds.plot_layout(layout_key='RNA_UMAP', color_by='RNA_cell_density')
time: 663 ms (started: 2021-08-22 18:57:14 +00:00)
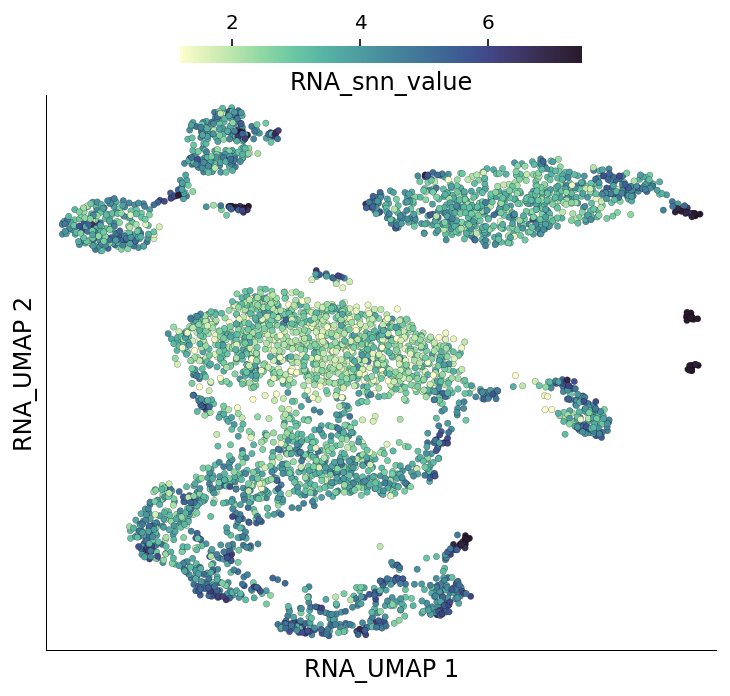
The dowsampling algorithm also identifies regions of the graph where cells form tightly connected groups by calculating mean shared nearest neighbours of each cell’s nieghbours. The tightly connected regions get a sampling award. These values can be accessed from under the cell metadata column RNA_snn_value.
[9]:
ds.plot_layout(layout_key='RNA_UMAP', color_by='RNA_snn_value')
time: 627 ms (started: 2021-08-22 18:57:15 +00:00)
That is all for this vignette.